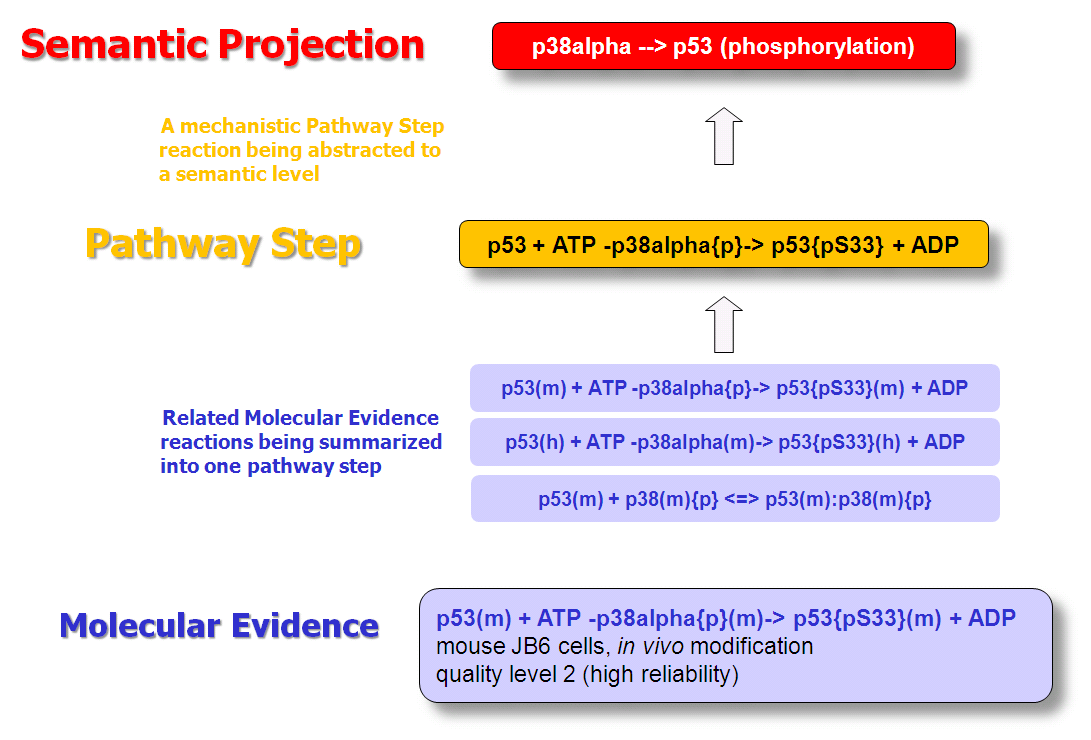
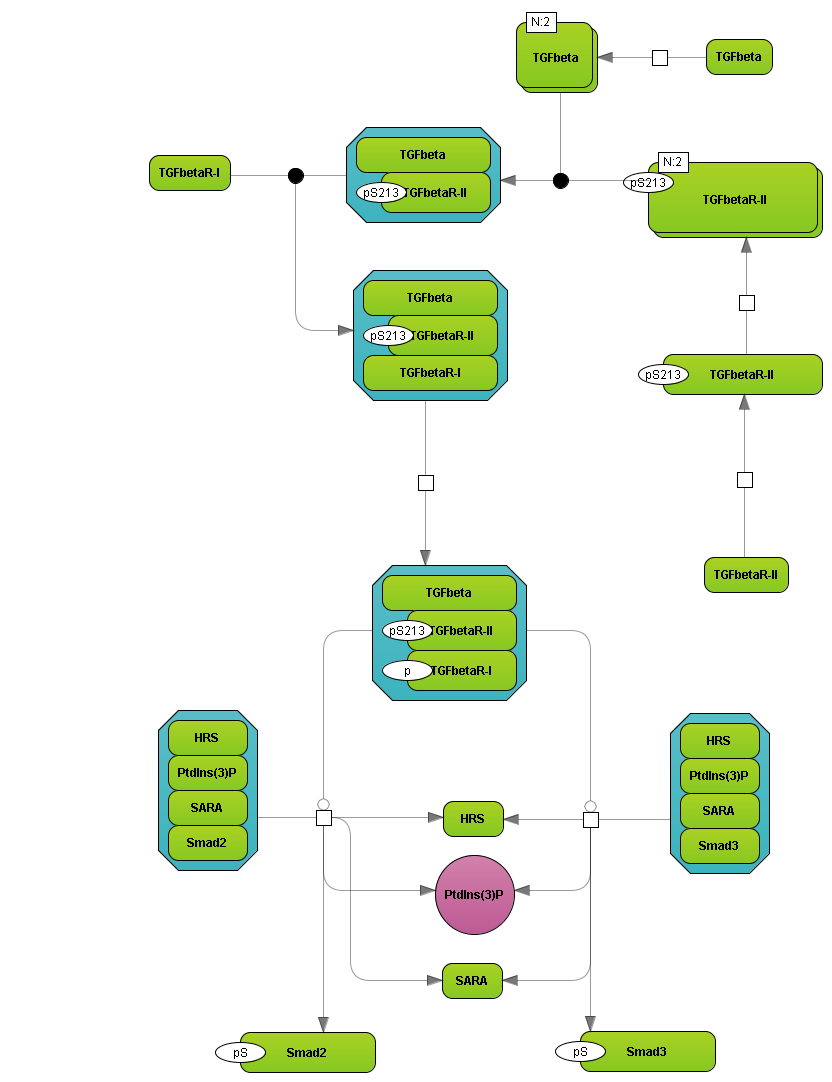
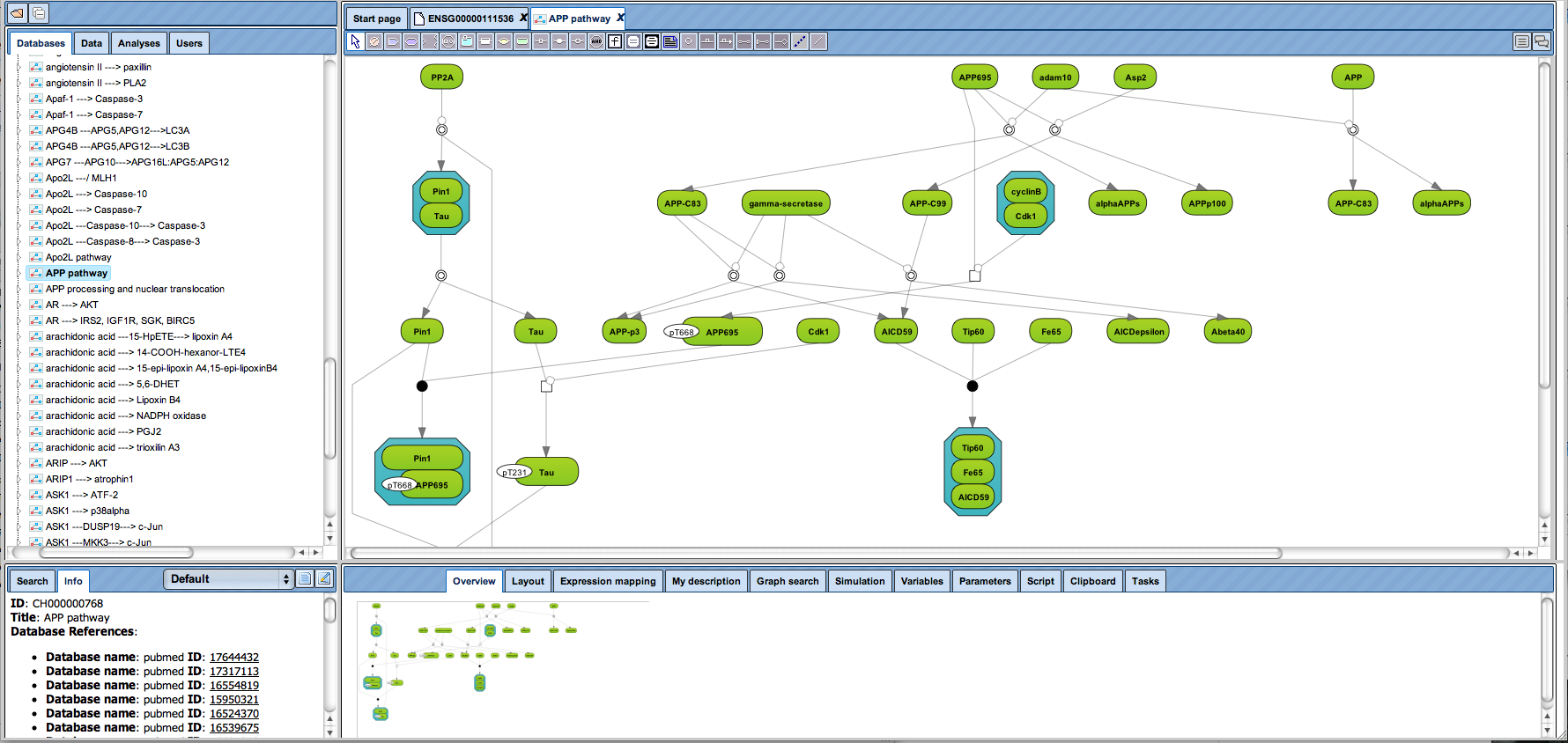
TRANSPATH® release 2023.2
The TRANSPATH® database on mammalian signal transduction and metabolic pathways contains these new data features:
· Increase in number of reactions
34,829 new binding reactions from recent publications between proteins in human, mouse, and rat have been added. In addition, 7,531 phosphorylation reactions by human receptor tyrosine kinases (RTK) were included.
· Update of links to Wikipathways and Reactome
Links from genes/proteins to the pathway databases Wikipathways (20230810) and Reactome (v85) have been updated.
Wingender, E., Hogan, J., Schacherer, F., Potapov, A.P., Kel-Margoulis, O. (2007) Integrating pathway data for systems pathology. In Silico Biol. 7:S17-S25. PubMed.
Kel, A., Voss, N., Jauregui, R., Kel-Margoulis, O., Wingender, E. (2006) Beyond microarrays: find key transcription factors controlling signal transduction pathways. BMC Bioinformatics 7:S13. PubMed.
Krull, M., Pistor, S., Voss, N., Kel, A., Reuter, I., Kronenberg, D., Michael, H., Schwarzer, K., Potapov, A., Choi, C., Kel-Margoulis, O., Wingender, E. (2006) TRANSPATH: an information resource for storing and visualizing signaling pathways and their pathological aberrations. Nucleic Acids Res. 34:D546-D551. PubMed
Choi, C., Crass, T., Kel, A., Kel-Margoulis, O., Krull, M., Pistor, S., Potapov, A., Voss, N., Wingender, E. (2004) Consistent re-modeling of signaling pathways and its implementation in the TRANSPATH database. Genome Inform. 15:244-254. PubMed
Choi, C., Krull, M., Kel, A., Kel-Margoulis, O., Pistor, S., Potapov, A., Voss, N., Wingender, E. (2004) TRANSPATH – a high quality database focused on signal transduction. Comp. Funct. Genomics 5:163-168. PubMed
Krull, M., Voss, N., Choi, C., Pistor, S., Potapov, A., Wingender, E. (2003) TRANSPATH: an integrated database on signal transduction and a tool for array analysis. Nucleic Acids Res. 31:97-100. PubMed
Schacherer, F., Choi, C., Götze, U., Krull, M., Pistor, S., Wingender, E. (2001) The TRANSPATH signal transduction database: a knowledge base on signal transduction networks. Bioinformatics 17:1053-1057. PubMed
Heinemeyer, T., Chen, X., Karas, H., Kel, A.E., Kel, O.V., Liebich, I., Meinhardt, T., Reuter, I., Schacherer, F., Wingender, E. (1999) Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 27:318-322. PubMed